Feb 8 , 2024 read
LLMs for relation extraction in clinical text
Clinical Insights: Leveraging LLMs for Relation Extraction in Clinical Text
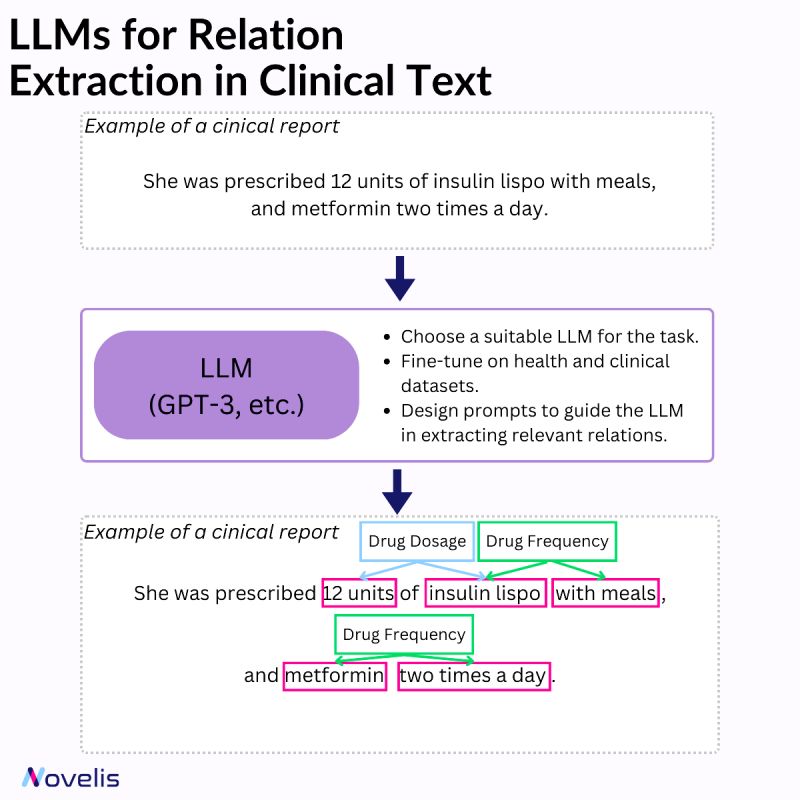
Relation extraction involves identifying connections between named entities in text. In the clinical domain, it helps extract valuable information from documents such as diseases, symptoms, treatments, and medications. Various techniques can be used for named entity recognition and relation extraction (rule-based systems, machine learning approaches, and hybrid systems that combine both).
Large Language Models (LLMs) have significantly impacted the field of machine learning, especially in natural language processing (NLP). These models, which are trained on large amounts of text data, are capable of understanding and generating natural language text with impressive accuracy. They have learned to identify complex patterns and semantic relationships within language, can handle various types of entities, and can be adapted to different domains and languages. They can also capture contextual information and dependencies more efficiently and are capable of transfer learning. When combined with a set of prompt-based heuristics and upon fine-tuning them on clinical data, they can be particularly useful for named entity recognition and relation extraction tasks.
Why is this essential? By identifying the relationships between different entities, it becomes possible to gain a better understanding of how various aspects of a patient’s health are connected. This, in turn, can help in developing effective interventions. For instance, clinical decision support can be improved by extracting relationships among diseases, symptoms, and treatments from electronic health records. Similarly, identifying potential interactions between different medications can ensure patient safety and optimize treatment plans. Automating the medical literature review process can facilitate quick access to relevant information.
Matching patients to clinical trials
Matching Patients to Clinical Trials Using Semantically Enriched Document Representation
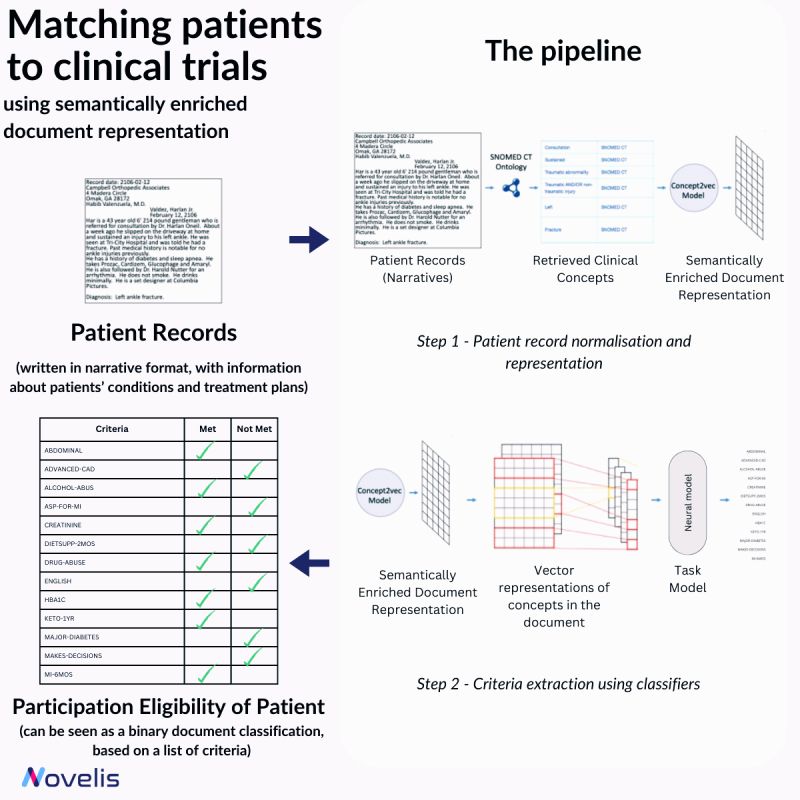
Recruiting eligible patients for clinical trials is crucial for advancing medical interventions. However, the current process is manual and takes a lot of time. Researchers ask themselves, “Which interventions lead to the best outcomes for a particular patient?” To answer this question, they explore scientific literature, match patients with potential trials, and analyze patient phenotypes to extract demographic and biomarker information from clinical notes. An approach presented in the paper “Matching Patients to Clinical Trials Using Semantically Enriched Document Representation” automates patient recruitment by identifying which patients meet the criteria for selection from a corpus of medical records.
This approach is utilized to extract important information from narrative clinical documents, gather evidence for eligibility decisions based on inclusion/exclusion criteria, and overcome challenges such as differences in reporting style with the help of semantic vector representations from domain ontologies. The SNOMED CT ontology is used to normalize the clinical documents, and the DBpedia articles are used to expand the concepts in SNOMED CT oncology. The team effectively overcame reporting style differences and sub-language challenges by enriching narrative clinical documents with domain ontological knowledge. The study involved comparing various models, and a neural-based method outperformed conventional machine learning models. The results showed an impressive overall F1-Score of 84% for 13 different eligibility criteria. This demonstrated that using semantically enriched documents was better than using original documents for cohort selection.
Why is this essential? This research is a significant step towards improving clinical trial recruitment processes. The automation of patient eligibility determination not only saves time but also opens avenues for more efficient drug development and medical research.
From AlphaFold to AlphaMissense
From AlphaFold to AlphaMissense: Models for Genetic Variations
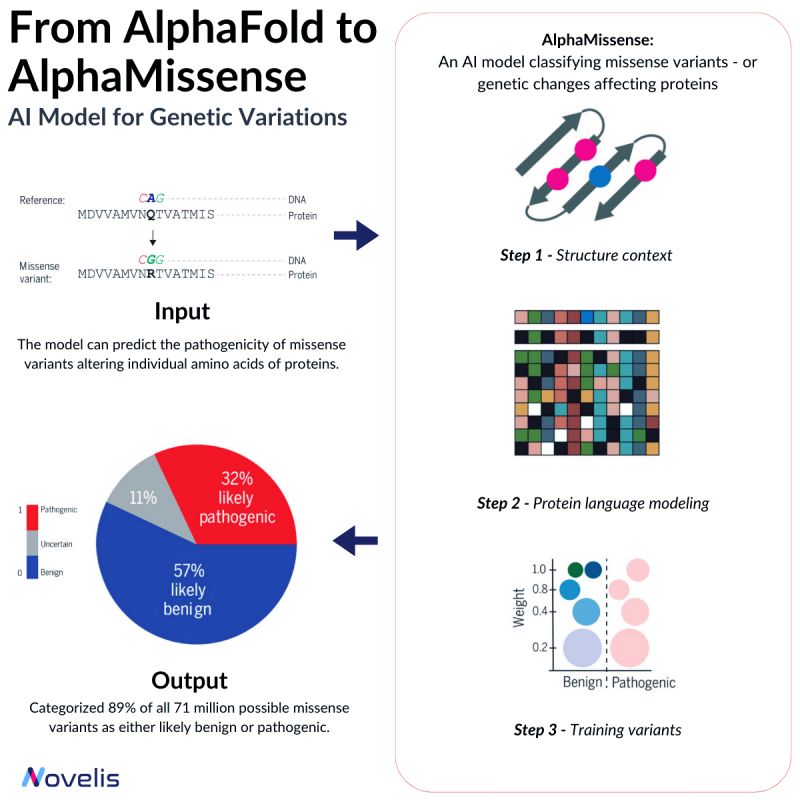
Missense mutations are responsible for contributing to a number of diseases, such as Marfan Syndrome and Huntington’s Disease.
These mutations cause a change in the sequence of amino acids in a protein, which can lead to unpredictable effects on the organism. Depending on their nature, missense mutations can either be pathogenic or benign.
Pathogenic variants significantly affect protein function, causing impairment in overall organism behavior, whereas benign variants have minimal or no effect on organism behavior.
Why is this essential? Despite identifying over 4 million missense variants in the human genome, only around 2% have been conclusively labeled as either pathogenic or benign.
The significance of the majority of missense variants is yet to be determined, making it difficult to predict their clinical implications. Hence, ongoing efforts aim to develop highly effective methods for accurately predicting the clinical implications of these variants.
The missense mutation problem shares similarities with the protein folding problem, both seeking to enhance explainability and predict outcomes related to variations in the amino acid structure.
In 2018, DeepMind and EMBL-EBI launched AlphaFold, a groundbreaking protein structures prediction model. Alphafold facilitates the prediction of protein structures from previously inaccessible amino acid sequences.
By leveraging the capabilities of Transfer Learning on binary labeled public databases (such as BFD, MGnify, and UniRef90), DeepMind proposes AlphaMissense, an AlphaFold finetune that achieves state-of-the-art predictions on ClinVar (a genetic mutation dataset) without the need for explicit training on such data.
The tool is currently available as a freely provided Variant Effect Predictor software plugin.
Introducing GatorTronGPT
Revolutionizing Healthcare Documentation: Introducing GatorTronGPT
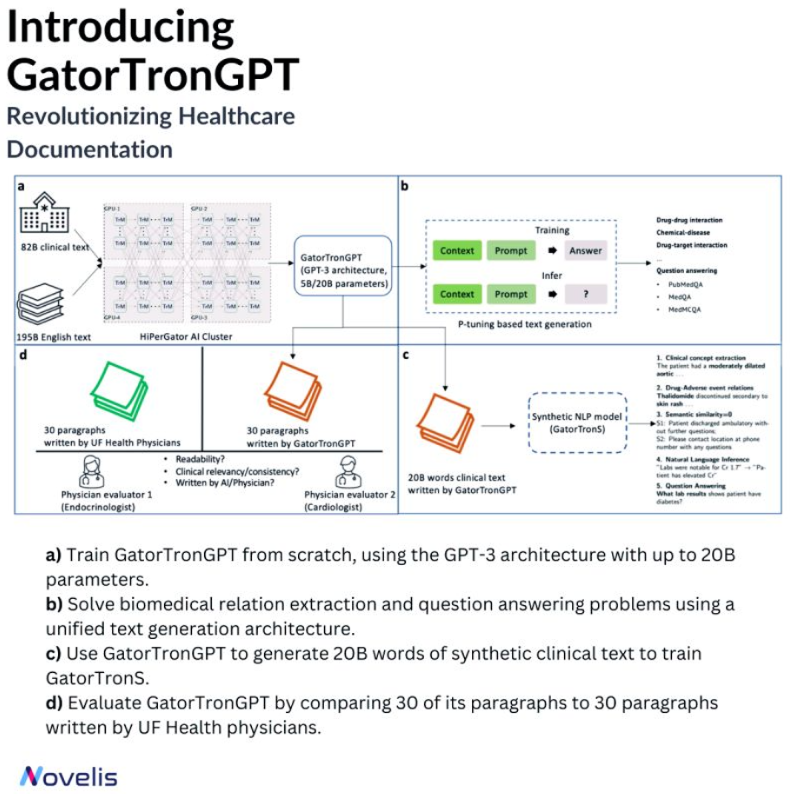
Meet GatorTronGPT, an advanced AI model developed by researchers at the University of Florida in collaboration with NVIDIA. This model transforms medical documentation, helping create precise notes. Its ability to understand complex medical language makes it a game-changer.
The language model was trained using the GPT-3 architecture. It was trained on a large amount of data, including de-identified clinical text from the University of Florida Health and diverse English text from the Pile dataset. GatorTronGPT was then employed to tackle two important biomedical natural language processing tasks: biomedical relation extraction and question answering.
A Turing test was conducted to evaluate the performance of GatorTronGPT. Here, the model generated synthetic clinical text paragraphs, and these were mixed with real-world paragraphs written by University of Florida Health physicians. The task was identifying which paragraphs were human-written and which were synthetic based on text quality, coherence, and relevance. Even experienced doctors could not differentiate between the generated and human-written paragraphs, which is a testament to the high quality of the GatorTronGPT output.
Powered by OpenAI’s GPT-3 framework, GatorTronGPT was trained on the supercomputer HiPerGator, with support from NVIDIA.
Why is this essential? By replicating the writing skills of human clinicians, GatorTronGPT allows healthcare professionals to save time, reduce burnout, and focus more on patient care.
